SCAN: Spatiotemporal Cloud Atlas for Neural cells
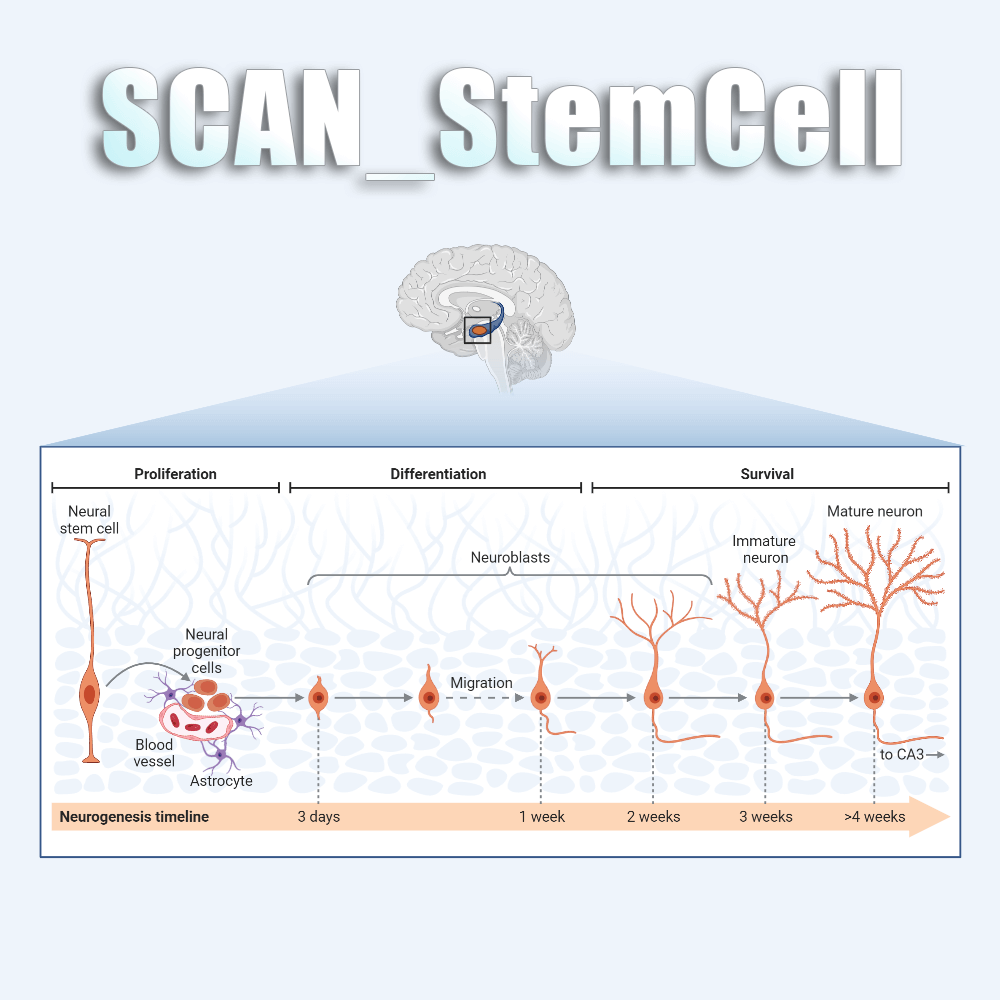
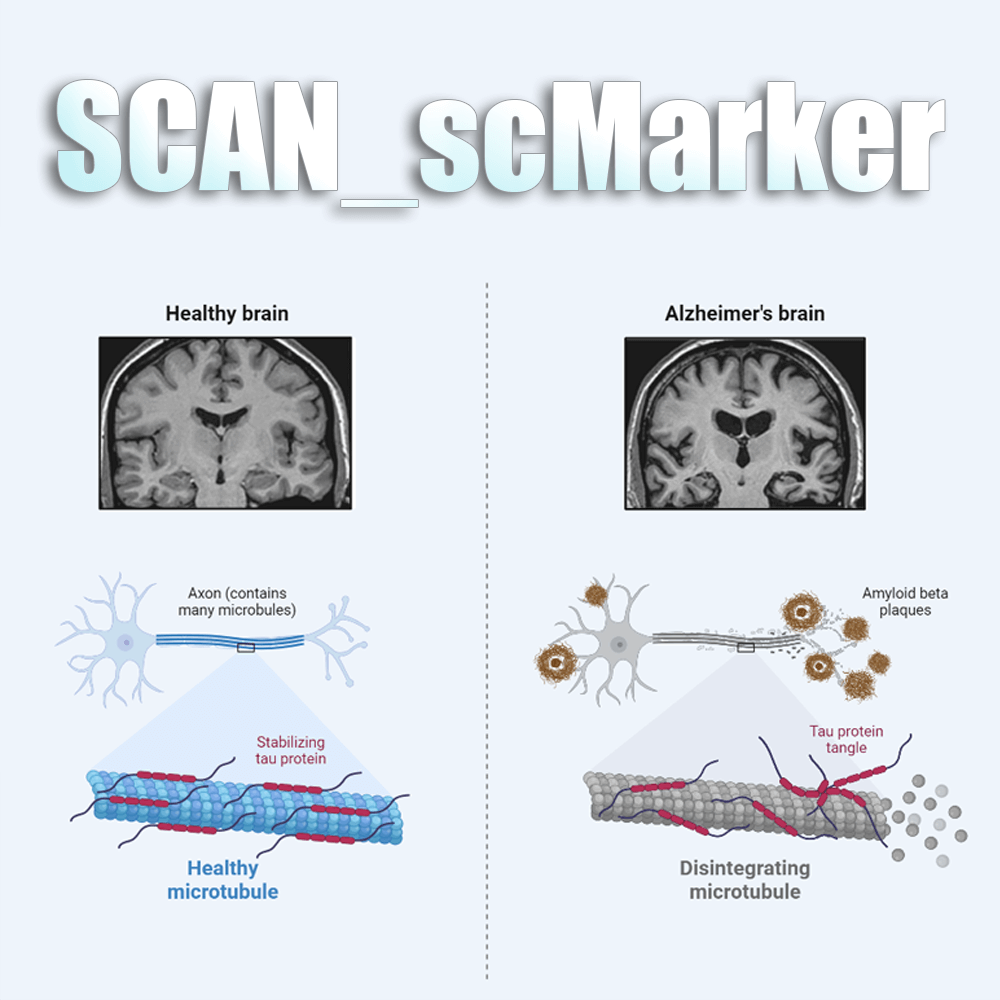
SCAN is the first neural database that combines single cell transcriptome and spatial transcriptome data, covering 10,679,684 cells from 960 species and 880 data sets generated by 26 single cell sequencing techniques. In addition, SCAN also covers 477 spatial transcriptome datasets including brain, retina, spinal cord, and embryos, and over 100 neurological diseases. On the other hand, SCAN also provides a wide range of analysis and visualization tools in the classification of neural cell types, evolution and development, tissue microenvironment, and disease development. It will enable users to comprehensively evaluate the composition of the nervous system in physiological and pathological conditions, and provide rich resources for research in the field of neuroscience.
The SCAN aims to better understand and utilize the currently known scRNA-seq and spatial transcriptomics datasets of the nervous system. Through a friendly user interface, researchers can freely obtain single cell and spatial transcriptomics data about the nervous system. Future published neural single cell sequencing and spatial transcriptomics data will be continuously updated in SCAN, and data generated by other multi-omics techniques in the nervous system may also be integrated into SCAN. In summary, SCAN will continue to enrich and expand the scientific community's full utilization of the valuable resource of single cell and spatial transcriptome maps of the nervous system.
960
Species
100
Diseases
20
Modules







Omics techniques selection